03:00
3 - What makes a model?
Survival analysis with tidymodels
Your turn
How do you fit a linear model in R?
How many different ways can you think of?
lmfor linear modelglmfor generalized linear model (e.g. logistic regression)glmnetfor regularized regressionkerasfor regression using TensorFlowstanfor Bayesian regressionsparkfor large data sets
To specify a model ![]()
- Choose a model
- Specify an engine
- Set the mode
To specify a model ![]()
![]()
To specify a model ![]()
![]()
To specify a model ![]()
To specify a model ![]()
![]()
All available models are listed at https://www.tidymodels.org/find/parsnip/
Your turn

Write the specification for a proportional hazards model.
Choose your engine.
All available models are listed at https://www.tidymodels.org/find/parsnip/
Extension/Challenge: Edit this code to use a different model. For example, try using a conditional inference tree as implemented in the partykit package - or try an entirely different model type!
03:00
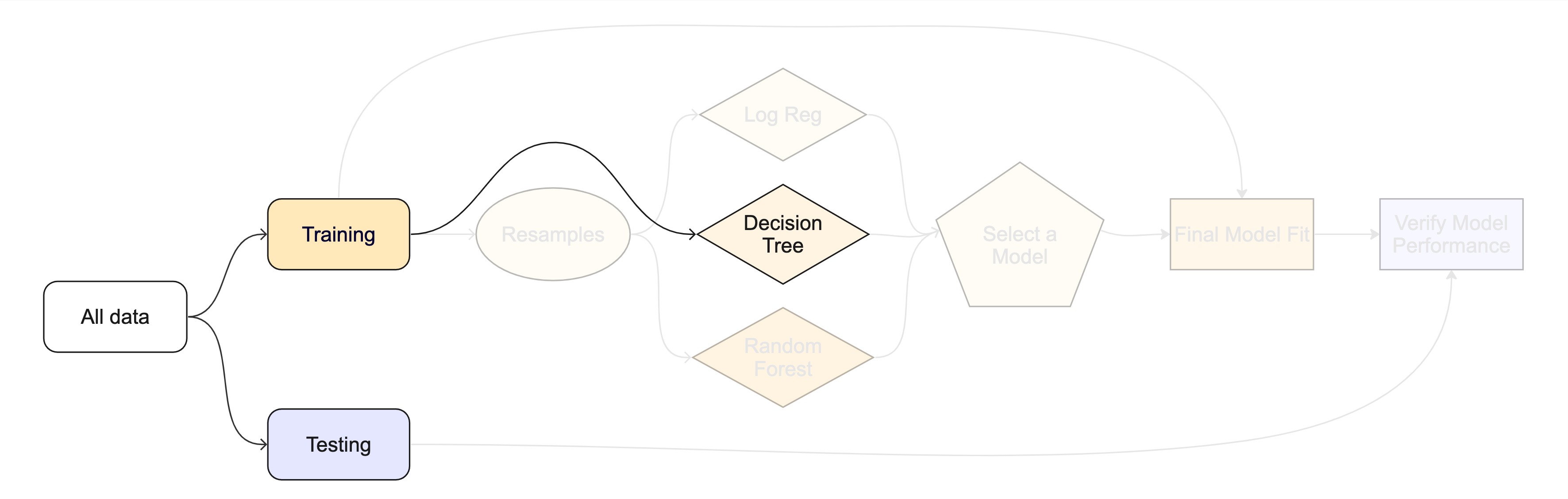
Models we’ll be using today
- Proportional hazards (PH) model
- Decision tree
Proportional hazards model
Proportional hazards model
- Hazard modeled via a baseline hazard and a linear combination of predictors:
\(\lambda(t | x_i) = \lambda_0(t) \cdot \exp (x_i^T \beta)\)
Proportional hazards model
- Hazard modeled via a baseline hazard and a linear combination of predictors:
\(\lambda(t | x_i) = \lambda_0(t) \cdot \exp (x_i^T \beta)\)
- The hazard is proportional over all time \(t\), and thus also the probability of survival is proportional.
Decision tree
Series of splits or if/then statements based on predictors
First the tree grows until some condition is met (maximum depth, no more data)
Then the tree is pruned to reduce its complexity
Decision tree
All models are wrong, but some are useful!
PH model
Decision tree
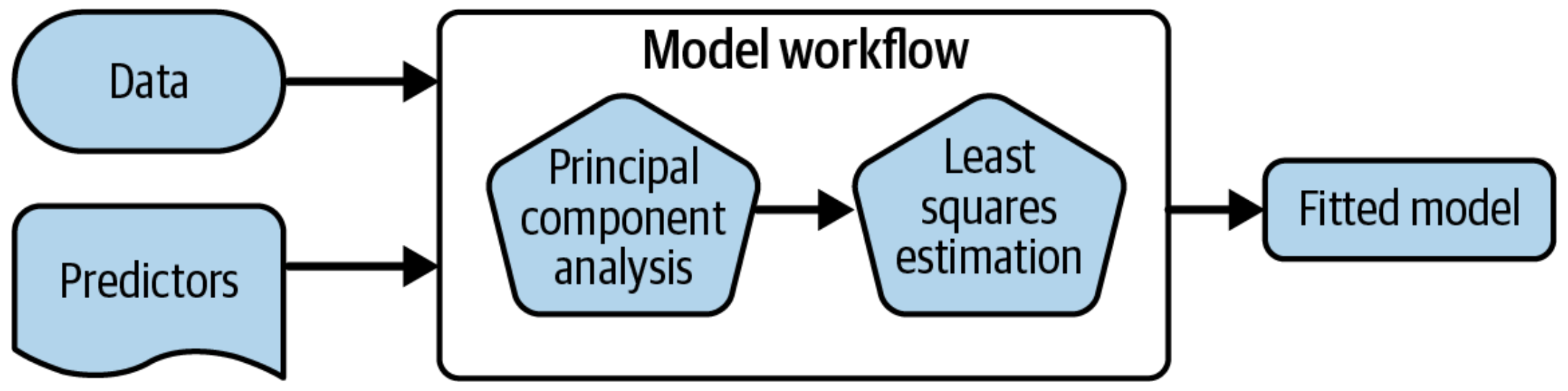
A model workflow
Workflows bind preprocessors and models
Fit a model spec ![]()
![]()
tree_spec <-
decision_tree() |>
set_mode("censored regression")
tree_spec |>
fit(event_time ~ ., data = cat_train)
#> parsnip model object
#>
#> $rpart
#> n= 1765
#>
#> node), split, n, deviance, yval
#> * denotes terminal node
#>
#> 1) root 1765 1981.0090 1.0000000
#> 2) neutered=no,unknown 696 576.4117 0.5937228 *
#> 3) neutered=yes 1069 1318.2450 1.1714650
#> 6) intake_condition=ill_mild,ill_moderatete,normal,under_age_or_weight,other 921 1063.5830 1.0808970 *
#> 7) intake_condition=feral,fractious 148 195.7649 2.3866130 *
#>
#> $survfit
#>
#> Call: prodlim::prodlim(formula = form, data = data)
#>
#> Right-censored response of a survival model
#>
#> No.Observations: 1765
#>
#> Pattern:
#> Freq
#> event 1116
#> right.censored 649
#>
#> $levels
#> [1] "0.593722848021764" "1.08089686322063" "2.38661313111009"
#>
#> attr(,"class")
#> [1] "pecRpart"Fit a model workflow ![]()
![]()
![]()
tree_spec <-
decision_tree() |>
set_mode("censored regression")
workflow() |>
add_formula(event_time ~ .) |>
add_model(tree_spec) |>
fit(data = cat_train)
#> ══ Workflow [trained] ════════════════════════════════════════════════
#> Preprocessor: Formula
#> Model: decision_tree()
#>
#> ── Preprocessor ──────────────────────────────────────────────────────
#> event_time ~ .
#>
#> ── Model ─────────────────────────────────────────────────────────────
#> $rpart
#> n= 1765
#>
#> node), split, n, deviance, yval
#> * denotes terminal node
#>
#> 1) root 1765 1981.0090 1.0000000
#> 2) neutered=no,unknown 696 576.4117 0.5937228 *
#> 3) neutered=yes 1069 1318.2450 1.1714650
#> 6) intake_condition=ill_mild,ill_moderatete,normal,under_age_or_weight,other 921 1063.5830 1.0808970 *
#> 7) intake_condition=feral,fractious 148 195.7649 2.3866130 *
#>
#> $survfit
#>
#> Call: prodlim::prodlim(formula = form, data = data)
#>
#> Right-censored response of a survival model
#>
#> No.Observations: 1765
#>
#> Pattern:
#> Freq
#> event 1116
#> right.censored 649
#>
#> $levels
#> [1] "0.593722848021763" "1.08089686322063" "2.38661313111009"
#>
#> attr(,"class")
#> [1] "pecRpart"Fit a model workflow ![]()
![]()
![]()
Your turn

Make a workflow with your own model of choice.
Extension/Challenge: Other than formulas, what kinds of preprocessors are supported?
03:00
Predict with your model ![]()
![]()
![]()
How do you use your new tree_fit model?
Predict with your model ![]()
![]()
![]()
How do you use your new tree_fit model?
Predict with your model ![]()
![]()
![]()
preds <- predict(tree_fit, new_data = demo_cats, type = "survival",
eval_time = seq(0, 365, by = 5))
preds
#> # A tibble: 50 × 1
#> .pred
#> <list>
#> 1 <tibble [74 × 2]>
#> 2 <tibble [74 × 2]>
#> 3 <tibble [74 × 2]>
#> 4 <tibble [74 × 2]>
#> 5 <tibble [74 × 2]>
#> 6 <tibble [74 × 2]>
#> 7 <tibble [74 × 2]>
#> 8 <tibble [74 × 2]>
#> 9 <tibble [74 × 2]>
#> 10 <tibble [74 × 2]>
#> # ℹ 40 more rowsPredict with your model ![]()
![]()
![]()
Predict with your model ![]()
![]()
![]()
Your turn

Run:
augment(tree_fit, new_data = demo_cats,
eval_time = seq(0, 365, by = 5))
What do you get?
03:00
The tidymodels prediction guarantee!
- The predictions will always be inside a tibble
- The column names and types are unsurprising and predictable
- The number of rows in
new_dataand the output are the same
The whole game - status update