demo_cat_preds <- augment(tree_fit, demo_cats, eval_time = ((1:10) * 30))
demo_cat_preds |> select(1:3)
#> # A tibble: 50 × 3
#> .pred .pred_time event_time
#> <list> <dbl> <Surv>
#> 1 <tibble [10 × 5]> 1.08 36
#> 2 <tibble [10 × 5]> 0.594 16+
#> 3 <tibble [10 × 5]> 0.594 12+
#> 4 <tibble [10 × 5]> 1.08 16
#> 5 <tibble [10 × 5]> 2.39 22
#> 6 <tibble [10 × 5]> 0.594 16
#> 7 <tibble [10 × 5]> 1.08 294
#> 8 <tibble [10 × 5]> 0.594 68+
#> 9 <tibble [10 × 5]> 1.08 31
#> 10 <tibble [10 × 5]> 0.594 24+
#> # ℹ 40 more rows4 - Evaluating models
Survival analysis with tidymodels
Predictions
For our demo data set of n = 50 cats, we’ll make predictions every 30 days for roughly the first 10 months:
Concordance
The concordance statistic (“c-index”, Harrell et al (1996)) is a metric that quantifies that the rank order of the times is consistent with the model score.
It takes into account censoring and does not depend on a specific evaluation time. The range of values is \([-1, 1]\).
Time-dependent metrics
Classification(ish) metrics
Most dynamic metrics convert the survival probabilities to events and non-events based on some probability threshold.
From there, we can apply existing classification metrics, such as
- Brier Score (for calibration)
- Area under the ROC curve (for separation)
More details on dynamics metrics at tidymodels.org
Converting to events
Converting to events
For a specific evaluation time point \(\tau\), we convert the observed event time to a binary event/non-event version, if possible.
\[ y_{i\tau} = \begin{cases} 0 & \text{if } \tau \lt t_{i} \text{ and } either \notag \\ 1 & \text{if } \tau \geq t_{i} \text{ and event} \notag \\ missing & \text{if } \tau \geq t_{i} \text{ and censored } \end{cases} \]
Dealing with missing outcome data
Without censored data points, this conversion would yield appropriate performance estimates since no event outcomes would be missing.
Otherwise, there is the potential for bias due to missingness.
We’ll use tools from causal inference to compensate by creating a propensity score that uses the probability of being censored/missing.
Case weights use the inverse of this probability. See Graf et al (1999).
Brier score
The Brier score is calibration metric originally meant for classification models:
\[ Brier = \frac{1}{N}\sum_{i=1}^N\sum_{k=1}^C (y_{ik} - \hat{\pi}_{ik})^2 \]
Brier score
For our application, we have two classes and case weights \(w_{it}\)
\[ Brier(t) = \frac{1}{N}\sum_{i=1}^N w_{it} \left[ \begin{aligned} &\underbrace{I(y_{it} = 0)((1 - y_{it}) - \hat{p}_{it})^2}_\text{non-events} \\ &+ \underbrace{I(y_{it} = 1)(y_{it} - (1 - \hat{p}_{it}))^2}_\text{events} \end{aligned} \right] \]
where \(N\) is the number of non-missing rows in the data.
Brier scores
demo_brier <- brier_survival(demo_cat_preds, truth = event_time, .pred)
demo_brier
#> # A tibble: 10 × 4
#> .metric .estimator .eval_time .estimate
#> <chr> <chr> <dbl> <dbl>
#> 1 brier_survival standard 30 0.151
#> 2 brier_survival standard 60 0.174
#> 3 brier_survival standard 90 0.148
#> 4 brier_survival standard 120 0.170
#> 5 brier_survival standard 150 0.159
#> 6 brier_survival standard 180 0.185
#> 7 brier_survival standard 210 0.147
#> 8 brier_survival standard 240 0.156
#> 9 brier_survival standard 270 0.162
#> 10 brier_survival standard 300 0.129Brier scores over evaluation time
Integrated Brier Score
Area Under the ROC Curve
We can use the standard ROC curve machinery on the indicators, probabilities, and censoring weights at evaluation time \(\tau\).
See Hung and Chiang (2010).
ROC curves measure the separation between events and non-events and are ignorant of how well-calibrated the probabilities are.
Area under the ROC curve
demo_roc_auc <- roc_auc_survival(demo_cat_preds, truth = event_time, .pred)
demo_roc_auc
#> # A tibble: 10 × 4
#> .metric .estimator .eval_time .estimate
#> <chr> <chr> <dbl> <dbl>
#> 1 roc_auc_survival standard 30 0.704
#> 2 roc_auc_survival standard 60 0.615
#> 3 roc_auc_survival standard 90 0.556
#> 4 roc_auc_survival standard 120 0.556
#> 5 roc_auc_survival standard 150 0.552
#> 6 roc_auc_survival standard 180 0.552
#> 7 roc_auc_survival standard 210 0.481
#> 8 roc_auc_survival standard 240 0.481
#> 9 roc_auc_survival standard 270 0.481
#> 10 roc_auc_survival standard 300 0.483ROC AUC over evaluation time
Setting evaluation times
- You can get predictions at any values of \(\tau\).
- During model development, we suggest picking a more focused set of evaluation times (for computational time).
- You should also pick a single time to perform your optimizations/comparisons with and list that value first in the vector. If 90 days was of particular interest over a 30-to-120-day span, you’d use
⚠️ DANGERS OF OVERFITTING ⚠️
Your turn
Why haven’t we done this?
05:00
The testing data are precious 💎
How can we use the training data to compare and evaluate different models? 🤔
Cross-validation ![]()
set.seed(123)
cat_folds <- vfold_cv(cat_train, v = 10) # v = 10 is default
cat_folds
#> # 10-fold cross-validation
#> # A tibble: 10 × 2
#> splits id
#> <list> <chr>
#> 1 <split [1588/177]> Fold01
#> 2 <split [1588/177]> Fold02
#> 3 <split [1588/177]> Fold03
#> 4 <split [1588/177]> Fold04
#> 5 <split [1588/177]> Fold05
#> 6 <split [1589/176]> Fold06
#> 7 <split [1589/176]> Fold07
#> 8 <split [1589/176]> Fold08
#> 9 <split [1589/176]> Fold09
#> 10 <split [1589/176]> Fold10Cross-validation ![]()
What is in this?
Set the seed when creating resamples
Bootstrapping ![]()
set.seed(3214)
bootstraps(cat_train)
#> # Bootstrap sampling
#> # A tibble: 25 × 2
#> splits id
#> <list> <chr>
#> 1 <split [1765/657]> Bootstrap01
#> 2 <split [1765/644]> Bootstrap02
#> 3 <split [1765/625]> Bootstrap03
#> 4 <split [1765/638]> Bootstrap04
#> 5 <split [1765/647]> Bootstrap05
#> 6 <split [1765/655]> Bootstrap06
#> 7 <split [1765/655]> Bootstrap07
#> 8 <split [1765/637]> Bootstrap08
#> 9 <split [1765/669]> Bootstrap09
#> 10 <split [1765/638]> Bootstrap10
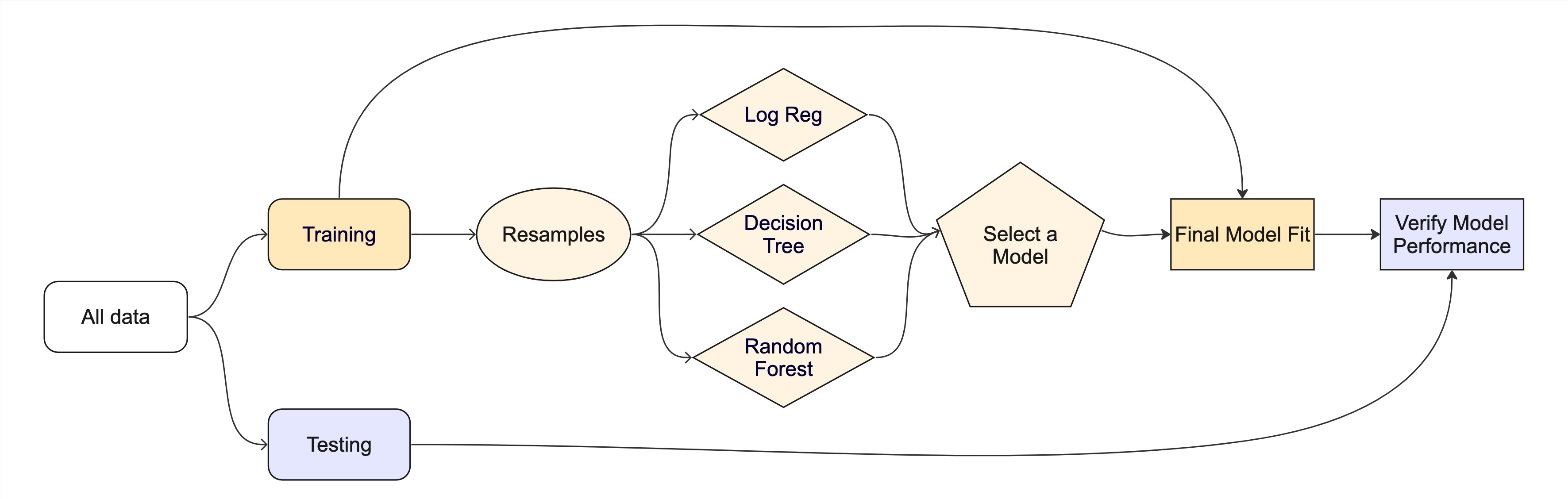
#> # ℹ 15 more rowsThe whole game - status update
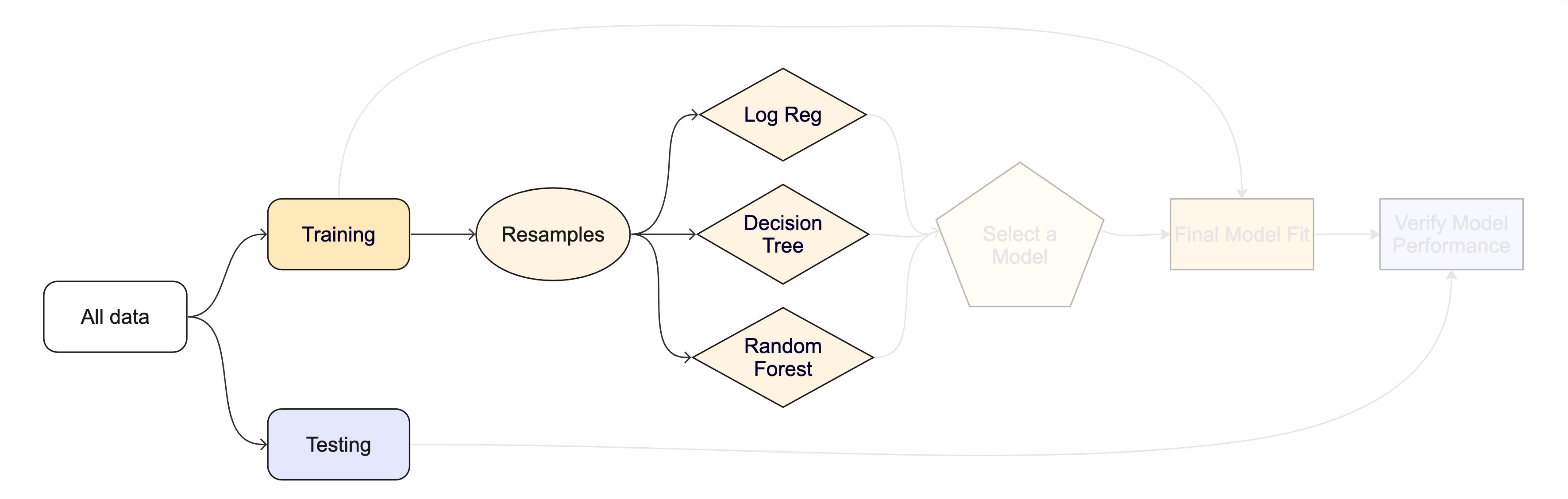
We are equipped with metrics and resamples!
Fit our model to the resamples
tree_wflow <- workflow(event_time ~ ., tree_spec)
tree_res <- fit_resamples(tree_wflow, cat_folds, eval_time = c(90, 30, 60, 120))
tree_res
#> # Resampling results
#> # 10-fold cross-validation
#> # A tibble: 10 × 4
#> splits id .metrics .notes
#> <list> <chr> <list> <list>
#> 1 <split [1588/177]> Fold01 <tibble [4 × 5]> <tibble [0 × 3]>
#> 2 <split [1588/177]> Fold02 <tibble [4 × 5]> <tibble [0 × 3]>
#> 3 <split [1588/177]> Fold03 <tibble [4 × 5]> <tibble [0 × 3]>
#> 4 <split [1588/177]> Fold04 <tibble [4 × 5]> <tibble [0 × 3]>
#> 5 <split [1588/177]> Fold05 <tibble [4 × 5]> <tibble [0 × 3]>
#> 6 <split [1589/176]> Fold06 <tibble [4 × 5]> <tibble [0 × 3]>
#> 7 <split [1589/176]> Fold07 <tibble [4 × 5]> <tibble [0 × 3]>
#> 8 <split [1589/176]> Fold08 <tibble [4 × 5]> <tibble [0 × 3]>
#> 9 <split [1589/176]> Fold09 <tibble [4 × 5]> <tibble [0 × 3]>
#> 10 <split [1589/176]> Fold10 <tibble [4 × 5]> <tibble [0 × 3]>Evaluating model performance ![]()
tree_res |>
collect_metrics()
#> # A tibble: 4 × 7
#> .metric .estimator .eval_time mean n std_err .config
#> <chr> <chr> <dbl> <dbl> <int> <dbl> <chr>
#> 1 brier_survival standard 30 0.266 10 0.00498 Preprocessor1_Model1
#> 2 brier_survival standard 60 0.264 10 0.00952 Preprocessor1_Model1
#> 3 brier_survival standard 90 0.330 10 0.00599 Preprocessor1_Model1
#> 4 brier_survival standard 120 0.155 10 0.00978 Preprocessor1_Model1We can reliably measure performance using only the training data 🎉
Where are the fitted models? ![]()
tree_res
#> # Resampling results
#> # 10-fold cross-validation
#> # A tibble: 10 × 4
#> splits id .metrics .notes
#> <list> <chr> <list> <list>
#> 1 <split [1588/177]> Fold01 <tibble [4 × 5]> <tibble [0 × 3]>
#> 2 <split [1588/177]> Fold02 <tibble [4 × 5]> <tibble [0 × 3]>
#> 3 <split [1588/177]> Fold03 <tibble [4 × 5]> <tibble [0 × 3]>
#> 4 <split [1588/177]> Fold04 <tibble [4 × 5]> <tibble [0 × 3]>
#> 5 <split [1588/177]> Fold05 <tibble [4 × 5]> <tibble [0 × 3]>
#> 6 <split [1589/176]> Fold06 <tibble [4 × 5]> <tibble [0 × 3]>
#> 7 <split [1589/176]> Fold07 <tibble [4 × 5]> <tibble [0 × 3]>
#> 8 <split [1589/176]> Fold08 <tibble [4 × 5]> <tibble [0 × 3]>
#> 9 <split [1589/176]> Fold09 <tibble [4 × 5]> <tibble [0 × 3]>
#> 10 <split [1589/176]> Fold10 <tibble [4 × 5]> <tibble [0 × 3]>🗑️
But it’s easy to save the predictions ![]()
# Save the assessment set results
ctrl_cat <- control_resamples(save_pred = TRUE)
tree_res <- fit_resamples(tree_wflow, cat_folds, eval_time = c(90, 30, 60, 120),
control = ctrl_cat)
tree_res
#> # Resampling results
#> # 10-fold cross-validation
#> # A tibble: 10 × 5
#> splits id .metrics .notes .predictions
#> <list> <chr> <list> <list> <list>
#> 1 <split [1588/177]> Fold01 <tibble [4 × 5]> <tibble [0 × 3]> <tibble>
#> 2 <split [1588/177]> Fold02 <tibble [4 × 5]> <tibble [0 × 3]> <tibble>
#> 3 <split [1588/177]> Fold03 <tibble [4 × 5]> <tibble [0 × 3]> <tibble>
#> 4 <split [1588/177]> Fold04 <tibble [4 × 5]> <tibble [0 × 3]> <tibble>
#> 5 <split [1588/177]> Fold05 <tibble [4 × 5]> <tibble [0 × 3]> <tibble>
#> 6 <split [1589/176]> Fold06 <tibble [4 × 5]> <tibble [0 × 3]> <tibble>
#> 7 <split [1589/176]> Fold07 <tibble [4 × 5]> <tibble [0 × 3]> <tibble>
#> 8 <split [1589/176]> Fold08 <tibble [4 × 5]> <tibble [0 × 3]> <tibble>
#> 9 <split [1589/176]> Fold09 <tibble [4 × 5]> <tibble [0 × 3]> <tibble>
#> 10 <split [1589/176]> Fold10 <tibble [4 × 5]> <tibble [0 × 3]> <tibble>But it’s easy to collect the predictions ![]()
tree_preds <- collect_predictions(tree_res)
tree_preds
#> # A tibble: 1,765 × 5
#> .pred id .row event_time .config
#> <list> <chr> <int> <Surv> <chr>
#> 1 <tibble [4 × 3]> Fold01 17 35 Preprocessor1_Model1
#> 2 <tibble [4 × 3]> Fold01 29 60 Preprocessor1_Model1
#> 3 <tibble [4 × 3]> Fold01 51 8 Preprocessor1_Model1
#> 4 <tibble [4 × 3]> Fold01 55 39+ Preprocessor1_Model1
#> 5 <tibble [4 × 3]> Fold01 62 36+ Preprocessor1_Model1
#> 6 <tibble [4 × 3]> Fold01 63 14 Preprocessor1_Model1
#> 7 <tibble [4 × 3]> Fold01 92 14+ Preprocessor1_Model1
#> 8 <tibble [4 × 3]> Fold01 95 37 Preprocessor1_Model1
#> 9 <tibble [4 × 3]> Fold01 96 19+ Preprocessor1_Model1
#> 10 <tibble [4 × 3]> Fold01 101 83 Preprocessor1_Model1
#> # ℹ 1,755 more rowsDecision tree 🌳
Random forest 🌳🌲🌴🌵🌴🌳🌳🌴🌲🌵🌴🌲🌳🌴🌳🌵🌵🌴🌲🌲🌳🌴🌳🌴🌲🌴🌵🌴🌲🌴🌵🌲🌵🌴🌲🌳🌴🌵🌳🌴🌳
Random forest 🌳🌲🌴🌵🌳🌳🌴🌲🌵🌴🌳🌵
Ensemble many decision tree models
All the trees vote! 🗳️
Bootstrap aggregating + random predictor sampling
- Often works well without tuning hyperparameters (more on this in a moment), as long as there are enough trees
Create a random forest model ![]()
Create a random forest model ![]()
rf_wflow <- workflow(event_time ~ ., rf_spec)
rf_wflow
#> ══ Workflow ══════════════════════════════════════════════════════════
#> Preprocessor: Formula
#> Model: rand_forest()
#>
#> ── Preprocessor ──────────────────────────────────────────────────────
#> event_time ~ .
#>
#> ── Model ─────────────────────────────────────────────────────────────
#> Random Forest Model Specification (censored regression)
#>
#> Main Arguments:
#> trees = 1000
#>
#> Computational engine: aorsfYour turn

Use fit_resamples() and rf_wflow to:
- keep predictions
- compute metrics
08:00
Evaluating model performance ![]()
ctrl_cat <- control_resamples(save_pred = TRUE)
# Random forest uses random numbers so set the seed first
set.seed(2)
rf_res <- fit_resamples(rf_wflow, cat_folds, eval_time = c(90, 30, 60, 120),
control = ctrl_cat)
collect_metrics(rf_res)
#> # A tibble: 4 × 7
#> .metric .estimator .eval_time mean n std_err .config
#> <chr> <chr> <dbl> <dbl> <int> <dbl> <chr>
#> 1 brier_survival standard 30 0.210 10 0.00455 Preprocessor1_Model1
#> 2 brier_survival standard 60 0.241 10 0.00796 Preprocessor1_Model1
#> 3 brier_survival standard 90 0.199 10 0.00960 Preprocessor1_Model1
#> 4 brier_survival standard 120 0.151 10 0.00840 Preprocessor1_Model1The whole game - status update
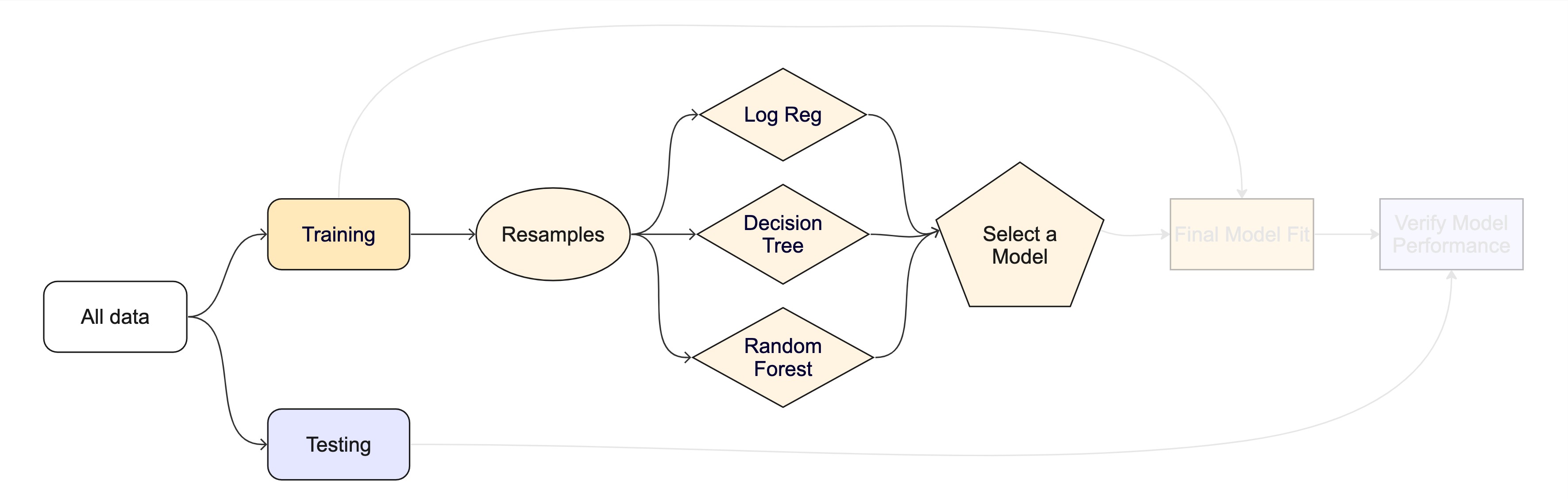
The final fit ![]()
Suppose that we are happy with our random forest model.
Let’s fit the model on the training set and verify our performance using the test set.
We’ve shown you fit() and predict() (+ augment()) but there is a shortcut:
# cat_split has train + test info
final_fit <- last_fit(rf_wflow, cat_split, eval_time = c(90, 30, 60, 120))
final_fit
#> # Resampling results
#> # Manual resampling
#> # A tibble: 1 × 6
#> splits id .metrics .notes .predictions .workflow
#> <list> <chr> <list> <list> <list> <list>
#> 1 <split [1765/442]> train/test split <tibble> <tibble> <tibble> <workflow>What is in final_fit? ![]()
collect_metrics(final_fit)
#> # A tibble: 4 × 5
#> .metric .estimator .eval_time .estimate .config
#> <chr> <chr> <dbl> <dbl> <chr>
#> 1 brier_survival standard 30 0.217 Preprocessor1_Model1
#> 2 brier_survival standard 60 0.225 Preprocessor1_Model1
#> 3 brier_survival standard 90 0.160 Preprocessor1_Model1
#> 4 brier_survival standard 120 0.108 Preprocessor1_Model1These are metrics computed with the test set
What is in final_fit? ![]()
extract_workflow(final_fit)
#> ══ Workflow [trained] ════════════════════════════════════════════════
#> Preprocessor: Formula
#> Model: rand_forest()
#>
#> ── Preprocessor ──────────────────────────────────────────────────────
#> event_time ~ .
#>
#> ── Model ─────────────────────────────────────────────────────────────
#> ---------- Oblique random survival forest
#>
#> Linear combinations: Accelerated Cox regression
#> N observations: 1765
#> N events: 1116
#> N trees: 1000
#> N predictors total: 18
#> N predictors per node: 6
#> Average leaves per tree: 142.739
#> Min observations in leaf: 5
#> Min events in leaf: 1
#> OOB stat value: 0.63
#> OOB stat type: Harrell's C-index
#> Variable importance: anova
#>
#> -----------------------------------------Use this for prediction on new data, like for deploying
The whole game